In this short post, I want to illustrate some graph clustering/partitioning ideas. First, I generate a random Gabriel graph with 50 vertices; how to generate such graphs I described in the previous post. First, we will need the following imports:
from geometric import random_gabriel_graph
import networkx as nx
import numpy as np
from gensim.models import Word2Vec
from sklearn.cluster import KMeans
import matplotlib.colors as mcolors
import matplotlib.pyplot as plt
And the code below gives us a graph.
G = random_gabriel_graph(50, seed=10)
points = nx.get_node_attributes(G, 'pos')
nx.draw_networkx_nodes(G, points, node_size=20)
nx.draw_networkx_edges(G, points, width=0.2)
plt.axis('off')
plt.savefig('gabrielGraph.png', dpi=300)

Node2Vec
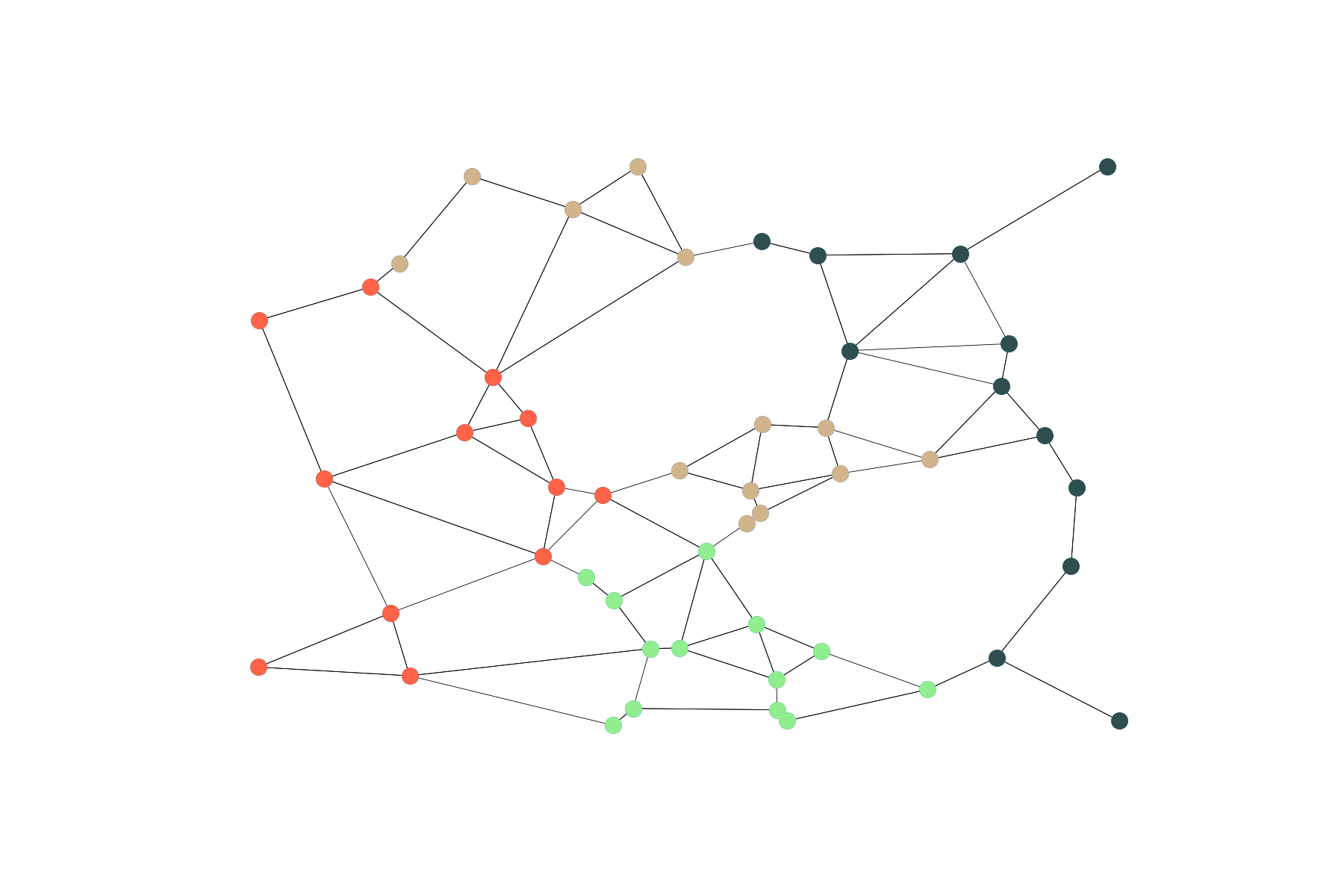
This one is my favorite; here we use a pre-trained Word2Vec model and random walks to extract features for every node. I like the idea due to its cross-disciplinarity, here we generate a large number of random walks on a graph and consider every walk as a sentence of some text. Finally, we use a pre-trained Word2Vec model from gensim to get a multidimensional vector for every node. The intuition is simple - if two words are often met in one sentence, their ‘meaning’ is correlated; analogously, if two nodes are often visited within one random walk, they are topologically ‘close’ and so should be their embeddings:
def sampleWalk(G, walkDepth, seed=None):
np.random.seed(seed)
walk = [np.random.choice(G.nodes())]
for _ in range(walkDepth-1):
walk.append(np.random.choice(list(G.neighbors(walk[-1]))))
return walk
def graphEmbed(G, walkNum=5000, walkDepth=7):
embedding = {}
sentences = []
for _ in range(walkNum):
sentences.append(sampleWalk(G, walkDepth))
model = Word2Vec(sentences, min_count=1)
for node in G.nodes():
embedding[node] = model.wv[node]
return embedding
Finally, let’s use the K-Means algorithm and cluster vertices of the graph, considered as points in 100-dimensional space according to their embedding:
def clusterWV(G, embedding, clusterNum = 4, seed=1):
np.random.seed(seed)
# fit
X = np.array(list(embedding.values()))
kmeans = KMeans(n_clusters=clusterNum, random_state=0).fit(X)
labels = kmeans.labels_
# draw
nodes = list(G.nodes())
pos = nx.get_node_attributes(G, 'pos')
nx.draw_networkx_edges(G, pos, width=0.2)
allColors = list(mcolors.CSS4_COLORS.values())
clusterColors = list(np.random.choice(allColors, clusterNum))
for label in list(range(clusterNum)):
labelCluster = [nodes[i] for i in range(len(nodes)) if labels[i] == label]
nx.draw_networkx_nodes(G, pos, nodelist=labelCluster, node_color=clusterColors[label], node_size=20)
What gives us the following partitioning.
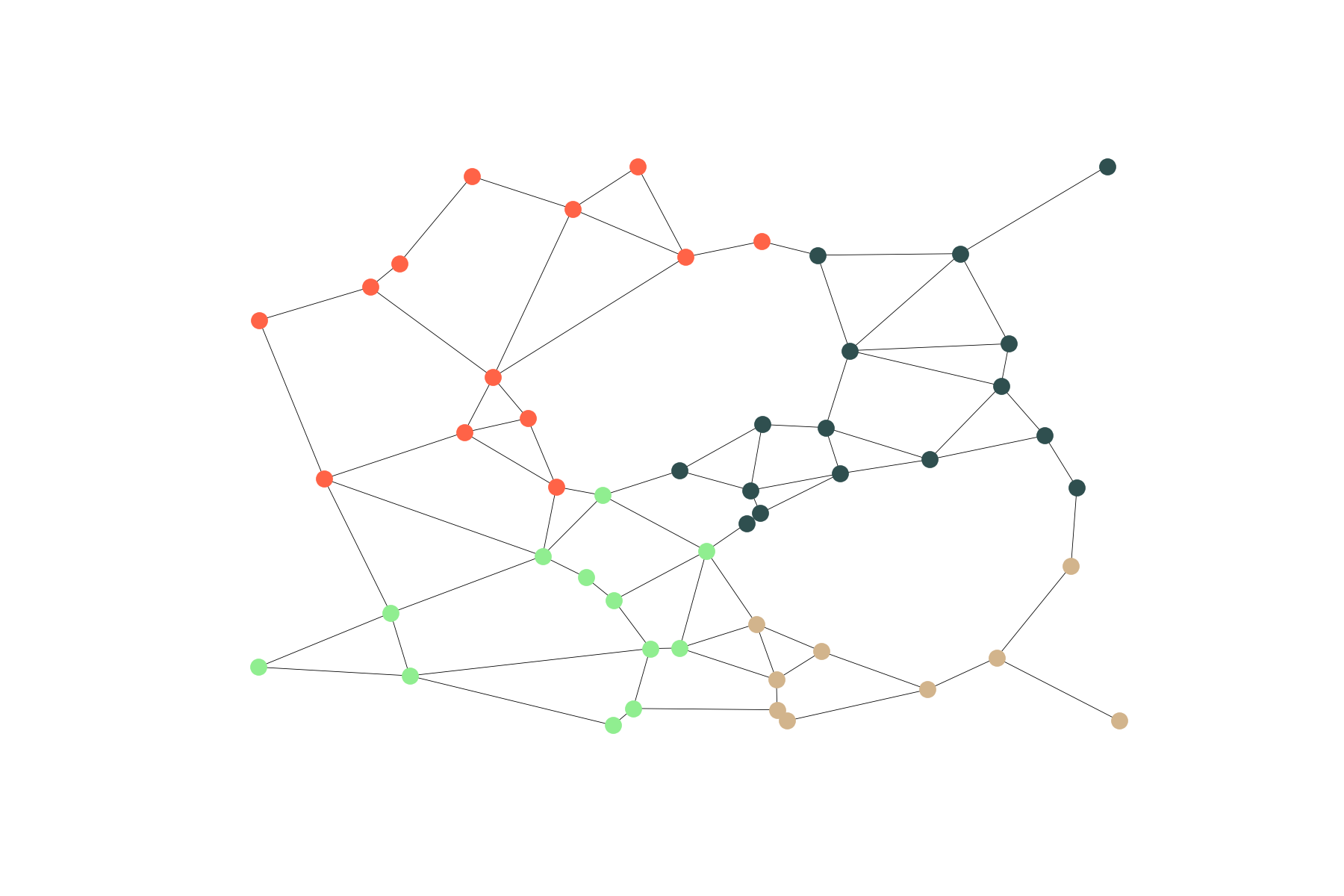
Geometrical
Gabriel graphs are naturally embedded in the Euclidean plane and we can use this embedding to cluster vertices with respect to its coordinates:
def clusterPOS(G, clusterNum = 4, seed=1):
np.random.seed(seed)
pos = nx.get_node_attributes(G, 'pos')
# fit
X = np.array(list(pos.values()))
kmeans = KMeans(n_clusters=clusterNum, random_state=0).fit(X)
labels = kmeans.labels_
# draw
nodes = list(G.nodes())
nx.draw_networkx_edges(G, pos, width=0.2)
allColors = list(mcolors.CSS4_COLORS.values())
clusterColors = list(np.random.choice(allColors, clusterNum))
for label in list(range(clusterNum)):
labelCluster = [nodes[i] for i in range(len(nodes)) if labels[i] == label]
nx.draw_networkx_nodes(G, pos, nodelist=labelCluster, node_color=clusterColors[label], node_size=20)
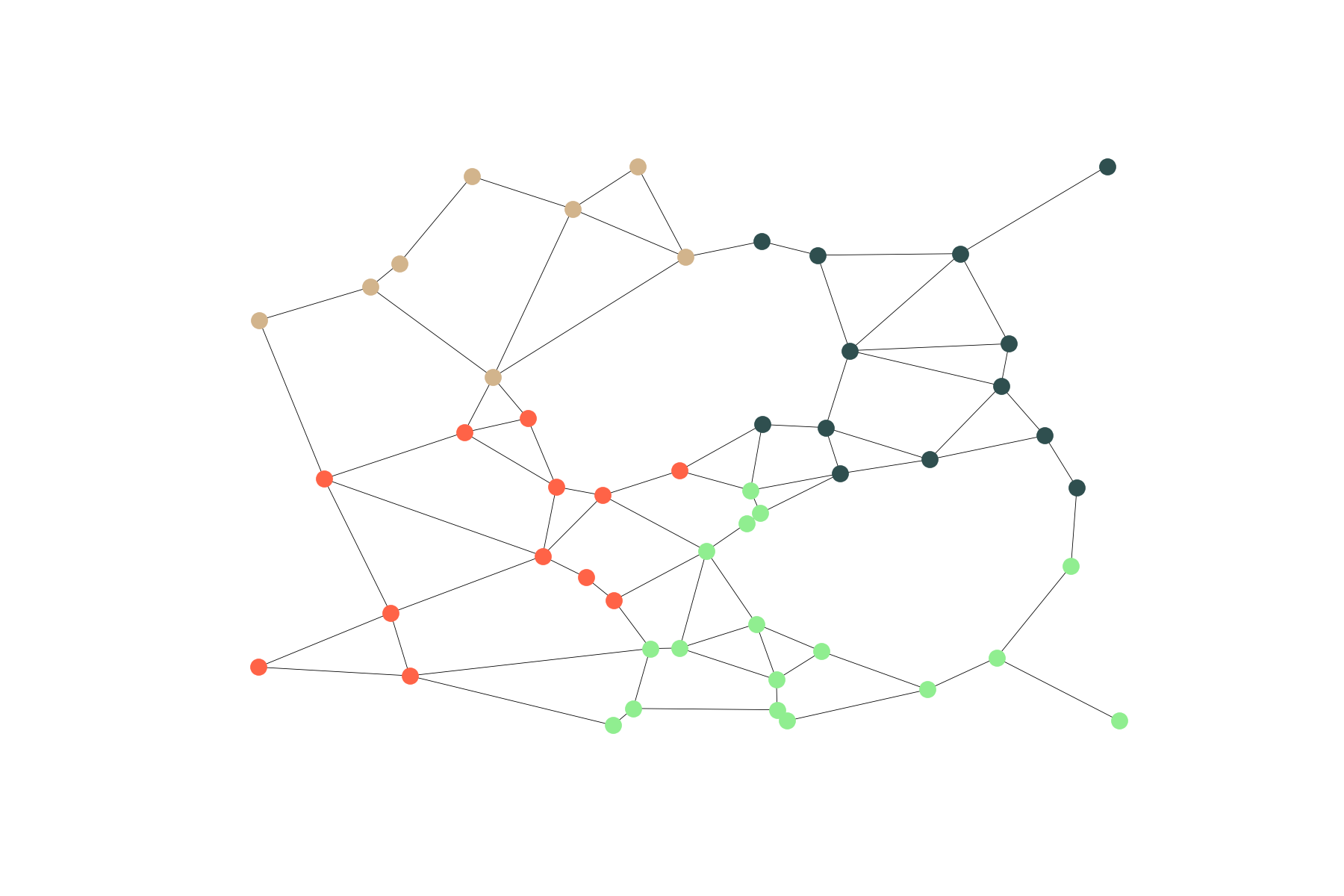
Kernighan–Lin Partitioning
Gabriel graphs are usually too sparse for Highly Connected Subgraph clustering. So I decided to use a more balanced graph partition that is still based on edge cuts:
def Bisec(G, clusterNum = 4):
from networkx.algorithms.community.kernighan_lin import kernighan_lin_bisection
H = G.copy()
components = list(sorted(nx.connected_components(H)))
while len(components) < clusterNum:
component1, component2 = kernighan_lin_bisection(H.subgraph(components.pop()).copy())
components = sorted(components + [component1, component2], key=len, reverse=False)
R = nx.empty_graph(len(G))
for c in components:
R = nx.compose(R, G.subgraph(c).copy())
return R